
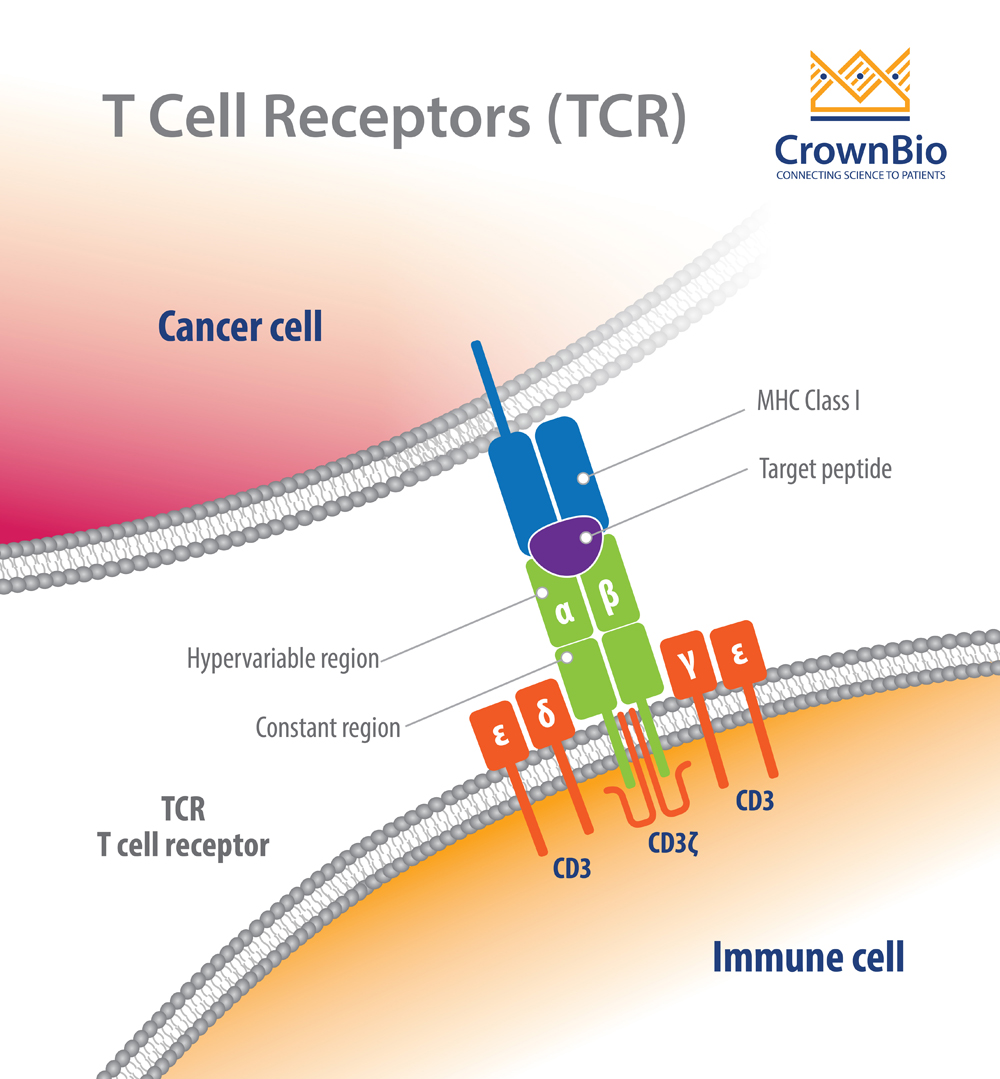
We identify the different sources of the observed variability, separating generation and selection effects. Using statistical models we learned the statistics of the processes generating this diversity from a large cohort of 651 individuals, including random generation and selection of T cells and their receptors. T cells express different surface receptors that can specifically bind molecules from viruses, bacteria or cancer cells.

The adaptive immune system is a naturally diverse set of many T cells with the potential to activate the organisms defense against specific threats. Known viral-specific TCRs follow the same generation and selection statistics as all TCRs. We find not only that most of the variability is driven by the VDJ generation process, but there is a large degree of consistency between individuals with the inter-individual variance of repertoires being about ∼2% of the intra-individual variance.
#T cell receptor repertoire analysis software#
We introduce a new software tool SONIA to facilitate inference of individual-specific computational models for the generation and selection of the TCR beta chain (TRB) from sequenced repertoires of 651 individuals, separating and quantifying the variability of the two processes of generation and selection in the population. However, the characterization of the variability is hampered by the limited size of the sampled repertoires. These processes lead to a large receptor variability within and between individuals. The diversity of T-cell receptor (TCR) repertoires is achieved by a combination of two intrinsically stochastic steps: random receptor generation by VDJ recombination, and selection based on the recognition of random self-peptides presented on the major histocompatibility complex.


 0 kommentar(er)
0 kommentar(er)
